Allozymic and chromosomal similarity in two Drosophila species
Abstract
D. setosimentum and ochrobasis are a pair of very close, partly sympatric species endemic to Hawaii island. Males of the two species differ strikingly in wing-pattern and there are altitudinal and breeding-site differences. Similarity indices have been calculated for both chromosomal (C) and allozymic (A) variants. Within the main populations of each species both kinds of data give coefficients above 0.98. Interspecific comparison of the main populations shows 0.66 (C) and 0.79 (A) An isolated population of ochrobasis from Kohala Volcans (Ohu), when compared with setosimentum, shows 0.68 (C) and 0.98 (A) Chromosomes are thus much more sensitive than allozymes in distinguishing these species; the same is true in the case of D. silvestris and heteroneura from the same forests. These morphologically distinct species, when compared, show 0,96 (A). All four species appear to be very new in the historical sense. In one area, about 2% of wild-caught D. setosimentum/ochrobasis are interspecific hybrids although adequate samples indicate that the separate gene pools have not broken down. The specific names should be retained but the two entities are perhaps best described as quite advanced semispecies in which reproductive isolation in nature is now nearly complete.
A revolution is brewing in contemporary systematics, especially at or near the level of species differences (1). Data on genetic variability within and between closely related populations can be used to establish highly sensitive indices of genetic similarity (2). A review of the biochemical variation within and between the members of the Drosophila willistoni group of species has recently appeared. A direct relationship between indices of genetic similarity and certain systematic designations was proposed (3).
If identity is taken as 1.0, local geographic populations of a species were shown to have mean genetic similarities of 0.97, subspecies and semispecies about 0.8, sibling species 0.52, and nonsibling species 0.35. The present paper reports genetic similarities within and between a pair of partially sympatric, very closely related species endemic to the Island of Hawaii. We introduce the use of indices based on chromosomal variability and compare them with indices based on allozymes. Biochemical similarity both within and between the species is high, although they are strongly differentiated morphologically and chromosomally. These species evidently are very new in the historical biological sense. We propose that their biochemical similarities are directly correlated with this circumstance.
Materials and methods
Specimens of Drosophila setosimentum and ochrobasis were collected in wet highland (1100-1600 m) forests on the “Big Island” of Hawaii, to which they are endemic. Except as supplemented below, details of geographical localities, sympatry, and cytological and electrophoretic techniques have been published (4). New collections were made at Puu Makaala, 8 km northeast of Olaa (locality 11, Fig. 1 in ref. 4) and at two sites along the western boundary between the Kau Forest Reserve and the Kahuku Ranch near the south end of the Island. One of these areas, called Kahuku Ranch - 3800 ft. (1150 m) yielded D. setosimentum. Other new collections, mostly D. ochrobasis, were made at Kipuka Pahipa, 7.2 km to the northeast of the Kahuku Ranch - 3800 ft. (1150 m) site. This is the same as locality 14, described in ref. 4.
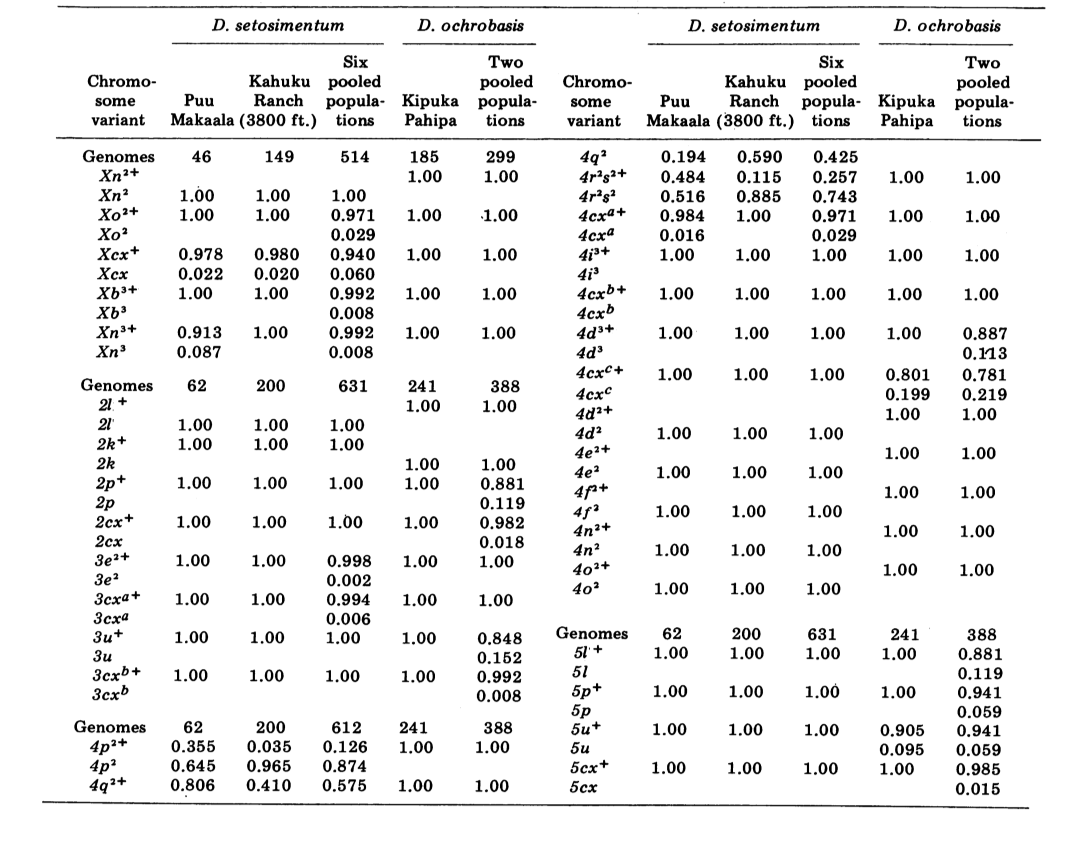
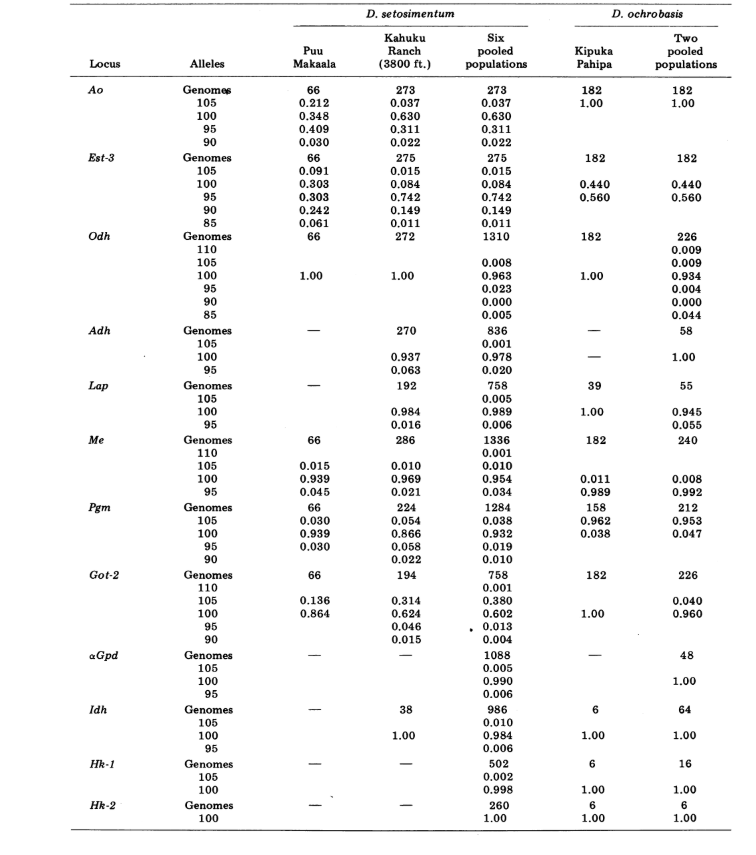
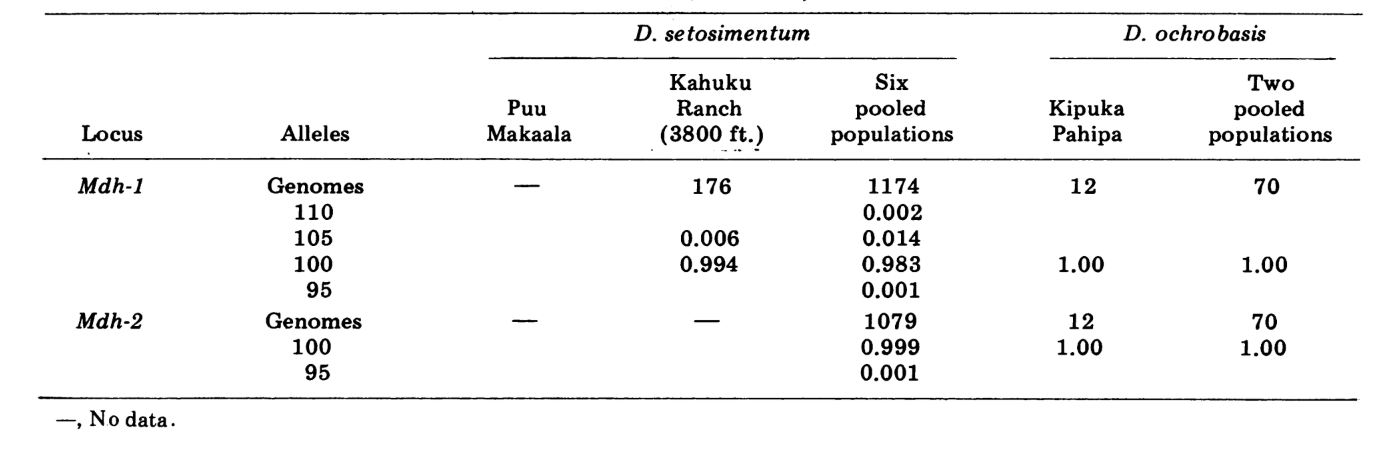
We report here data on three additional allozyme loci, aldehyde oxidase (Ao), a β-napthyl acetate esterase locus (Est-3) and an additional hexokinase (Hk-2), making a maximum of fourteen used for comparison of populations. The two species show variation at a total of 30 polytene chromosome sections due to inversions or complex chromosomes. Each section was scored in each salivary gland chromosome smear examined; that is, it was recorded as heterozygous for the variant, homozygous for the variant, or homozygous for the standard condition. These data have been used to calculate similarity coefficients in the same manner as for electrophoretic data. Table 2 lists a new inversion, Xn3, from Puu Makaala. Reexamination of old slides from Mawae, Kipuka-4400 (1340 m) ft. and Olaa show its frequency in these populations to be 0.056, 0.032, and 0.005, respectively.
Observations
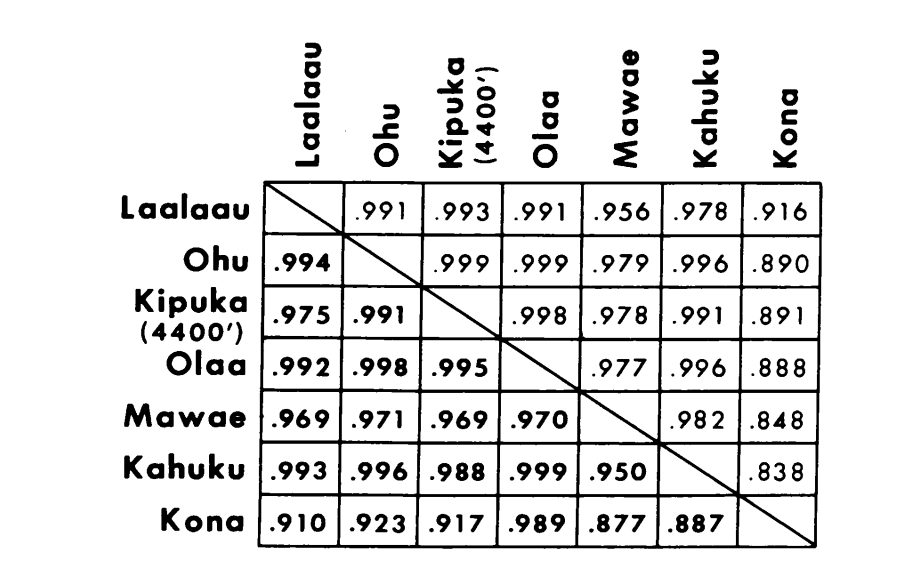
The new data and those from (4) were used to calculate similarity coefficients (Nei's I) for both allozymes (7-14 loci per comparison) and chromosomes (30 sections per comparison), for seven populations of D. setosimentum and three of ochrobasis (Tables 1-3 and Fig. 1).
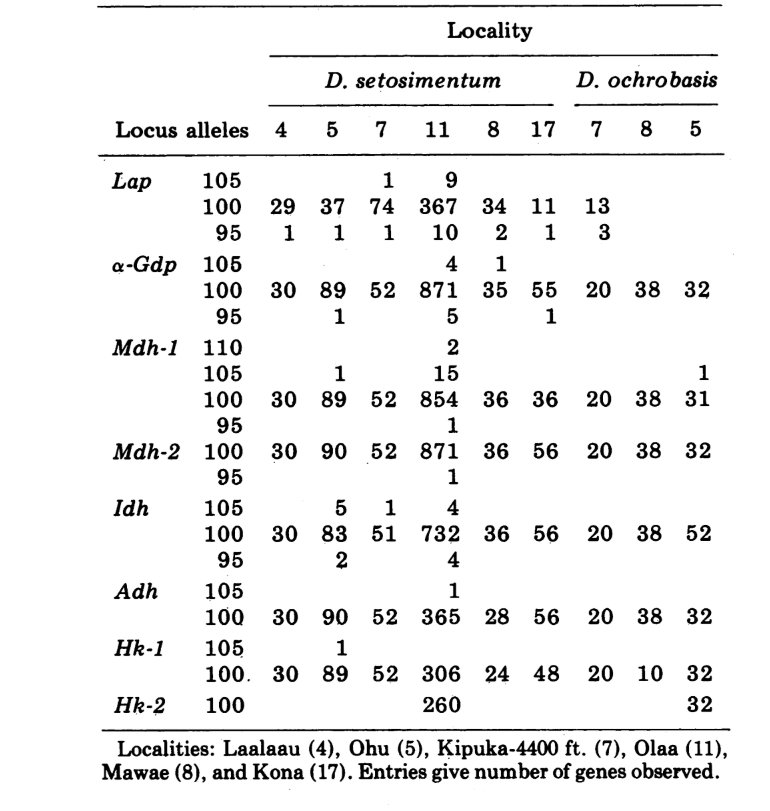
The first six populations of D. setosimentum listed in Fig. 1 are all from the highlands of the windward side of the island. They show uniformly high coefficients for both chromosomes and allozymes and may be pooled. Mean I with standard error is given on the top line in the body of Table 4. Most comparisons show some difference from the Kona population, especially chromosomally (Fig. 1 and Table 4). The latter is geographically isolated from the others on the southwest side of the Island and displays a fixed metaphase heterochromatin difference which is not included in the calculation of the polytene chromosome coefficients.
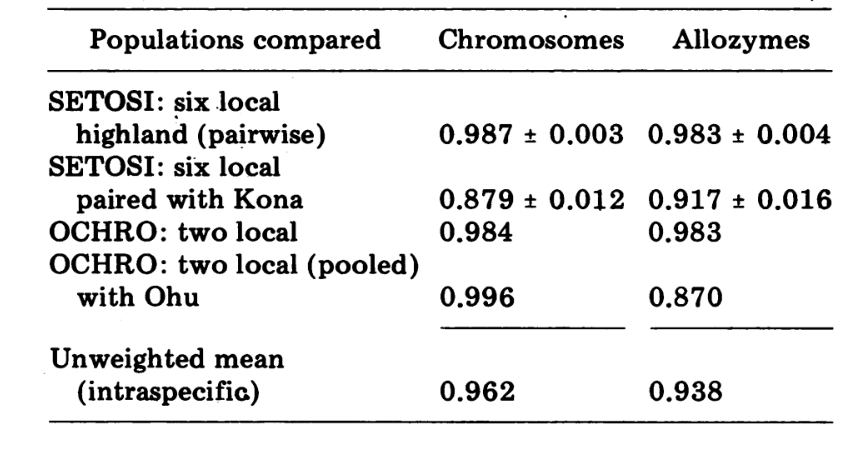
The data from D. ochrobasis were handled in a similar manner (Tables 1-5). Pairwise examination of the two populations, Kipuka Pahipa and the combined Mawae-Kipuka 4400 ft. samples show high similarity (Table 4). When pooled, these samples show a small difference electrophoretically from the ochrobasis population of Ohu, which is isolated from the others on the Kohala volcano in the northwest corner of the Island. The overall unweighted intraspecific means, however, are both quite high and quite similar (Table 4).
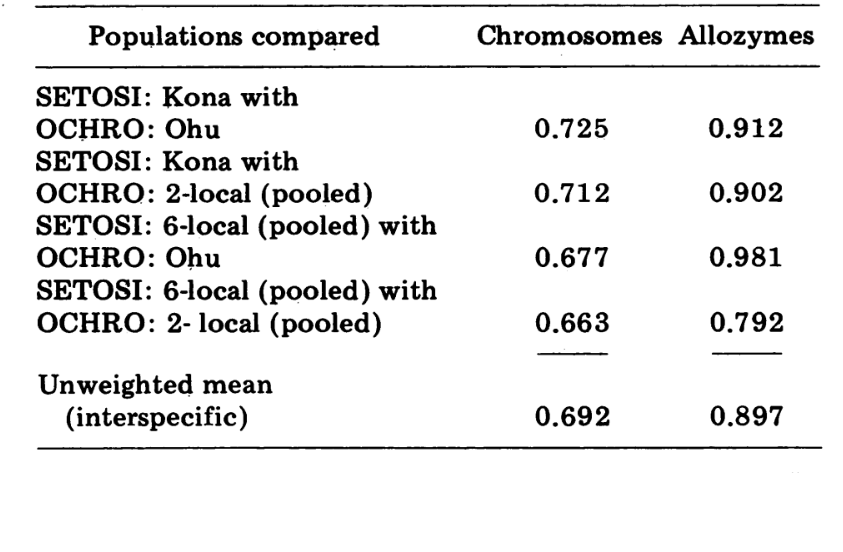
Interspecific comparisons are shown in Table 5. The comparisons in the Table are listed in order of decreasing chromosomal similarity. The data show that the interspecific chromosomal comparisons are much more efficient in distinguishing the species than are those based on allozymes; in fact, three of the four latter comparisons are above 0.9. The case of the Ohu population of ochrobasis is interesting. Although it stands out as different when allozymes within its own species are compared (0.870, Table 4), it shows extraordinary allozymic similarity to setosimentum (0.981 with the highland pool and 0.912 with Kona, Table 5).
Discussion
Although cytological differentiation between these two species is considerable, biochemical differences are small. The two entities, however, do not appear to be subspecies. Individuals of each may be clearly recognized in most sympatric situations by any one of an extensive series of fixed inversions and there are altitudinal (4) and breeding-site differences (5) as well. Males of the two species differ strikingly in color pattern on the wings (6).
On the other hand, there is considerable evidence to suggest that D. setosimentum and ochrobasis might be considered semispecies (7). The inference that some hybridization has occurred between them in the past (4) has now been confirmed by a recent discovery (8). In the Kahuku Raneh-Kau Forest Reserve area near the south end of the Island, one naturally-occurring Fl and three backeross hybrids between tween the species have been unequivocally recognized. This amounts to about two per cent of the wild flies examined in this area. In view of the substantial natural samples studied and the wealth of both electrophoretic (9) and cytological markers in that area, the interbreeding can be judged to be rather sparse.
Secondary sexual differences, such as those that characterize the males of these species, do not militate against the proposal of semispecies status. Indeed, such character distinguish some of the classical cases of semispeeies, as, for example, in certain birds of paradise (7). As in the birds, these secondary sexual characters are concerned with the complex territorial (lek) and sexual behavior manifested by many of the “picture-winged” group of Hawaiian Drosophila to which these flies belong (10).
Finally, the most inclusive electrophoretic comparison (4th comparison, Table 5) gives a value of 0.792, close to that proposed for the semispecies of Drosophila paulistorum (3). Unlike the latter, however, D. setosimentum and ochrobasis appear to be best described as quite advanced semispecies, in which reproductive isolation in nature is still incomplete. We favor the retention of the two specific names for the two entities.
There remains, however, the interesting fact that certain populations of the two species are virtually indistinguishable electrophoretically. The most extreme of these is the Ohu population of ochrobasis, which shows a similarity index of 0.981 when compared with D. setosimentum. The parallel cytological information on this population precludes the possibility that the electrophoretic similarity is due to current hybridization.
Close interspecific electrophoretic similarity is not unknown between other species of Drosophila from the Island of Hawaii (11). For example, the sympatric species D. silvestris and D. heteroneura have extensive morphological (12) and cytological (13, 14) differences. They show ethological isolation (15) as well as other biological differences (16). All this implies considerable genetic difference in addition to the inversions. Yet these two species show an allozymic similarity coefficient of 0.96 (11). According to the proposals in (3), this would place them as local populations of the same species, an untenable interpretation.
Hawaii Island is geologically new (less than 7 x 105 years). Species endemic to the Island are necessarily even newer. One of us (17) has suggested that speciation in these and other forms may have involved a forced genetic reorganization following the stochastic effects of an allopatric founder event. Unlike the subjects of this paper, however, quite a few instances are known in whieh species are formed without any detectable polytene chromosome reorganization. One cluster of eight (now 9) homosequential species related to D. grimshawi is known (18). In like manner, the present cases show that species can indeed also be formed with only a small amount of allozymic reorganization. We suggest that this is related to the recency of the speciational events. Possibly the two populations have just recently emerged from a strongly stochastic stage in their life history as species. This stage may be characterized principally by recombinational reorganization relative to the ancestral species and the establishment of new internally balanced genetic systems. The phase of new adaptational response and environmental tracking may have only just begun.
This view also emphasizes a point recently made in a discussion of the rather striking biochemical similarities of man and chimpanzee (19). Speciational events may involve regulatory genes which are not normally revealed by current methods in population genetics.
Authors: H. L. Carson, W. E. Johnson, P. S. Nair, F. M. Sene

/public-service/media/default/427/kGS1A_671a139147732.png)
/public-service/media/default/430/DxsI5_671a145105a71.jpeg)
/public-service/media/default/431/gortG_671a1464675a1.jpeg)