Genetic Variation In Hawaiian Drosophila. Iv. Allozymic Similarity Between D. Silvestris And D. Heteroneura From The Island Of Hawaii
Abstract
The species are endemic to the newest island in the archipelago and are broadly sympatric. They are easily distinguished morphologically in both sexes. Using standard electrophoretic procedures, we have examined 25 loci encoding for structural proteins from 539 silvestris and 325 heteroneura collected at three widely-separated localities where the two species are sympatric. Pairwise comparisons within and between the species show the following coefficients of genetic identity (Nei's I): within silvestris 0.961 ± 0.01; within heteroneura 0.949 ± 0.02; between silvestris and heteroneura, 0.939 ± 0.01. Neither the differences within nor between the species are significant. There are no fixed allelic differences either within or between the species. At the three areas of sympatry, the species show gene frequency differences P < 0.05) at 9, 11 and 13 loci respectively. This is not much different from the variation within either one of the species across the three localities. The two species have similar heterozygosity (H) levels (silvestris, 0.083; heteroneura, 0.089) and percent of polymorphic loci (both 0.37). It is suggested that despite their morphological divergence, these species are much more newly formed than classical sibling species. Significant allozymic differences may not have had time to accumulate.
DROSOPHILA silvestris and D. heteroneura are a pair ofclosely related giant species endemic only to the rain forests of Hawaii island, the newest in the archipelago. They are sympatric in most areas except at higher altitudes where populations tend to be exclusively silvestris.
The species are easily distinguished morphologically (Hardy 1965) not only by the extraordinary wide head in both sexes of heteroneura but also by the color patterns of the face, thorax, pleurae and abdomen and by differences in the pattern of wing markings. D. silvestris carries six unique inversions in polymorphic state; a seventh inversion is polymorphic Within both species (Carson and Stalker 1968; Craddock and Carson 1975). There are no fixed inversions between the species, and the two share a common set of standard sequences, i.e., are chromosomally homosequential.
Although there is strong sexual isolation between the species in laboratory crosses (Ahearn et al. 1974; Kaneshiro 1976) F1, hybrids have been obtained from both reciprocal crosses (Craddok 1974; Ahearn and Val 1975). Both sexes of these F1, hybrids are fully fertile and genetic segregation of the morphological species differences has been observed in the F, generation and in backcrosses (Val 1976).
Preliminary studies of allozymes have shown extraordinary similarity between the two species (S = 0.96; Johnson et al. 1975; Johnson and Carson 1975). The present paper extends the study of genetic variability within and between the two species to soluble proteins encoded by 25 loci. The data are from three areas of sympatry. The results confirm the striking biochemical similarity of the species despite the genetic differences between them in morphology, inversions and behavior.
Materials and methods
In 1974 and 1975, we collected the two species from three areas of moderate elevation on Hawaii (The Big Island) where they are sympatric. These are (Figure 1): (1) Kahuku Ranch in the southern part of the Kau District, (elevation 1158 m); (2) Olaa, forest near the Volcano Experiment Station on Wright Road, Puna District (elevation 1238 m) and (3) Pauahi, South Kona District (elevation 1311 m). Newly captured males were used for electrophoresis as soon as possible after capture. Females were isolated and allowed to produce F1 progeny for use in salivary gland preparations. After producing larvae, the mothers were used for electrophoresis. Only the abdomen of the flies was subjected to electrophoresis; the remainder (head, thorax and appendages) was mounted and placed in a collection to be used for morphological analyses.
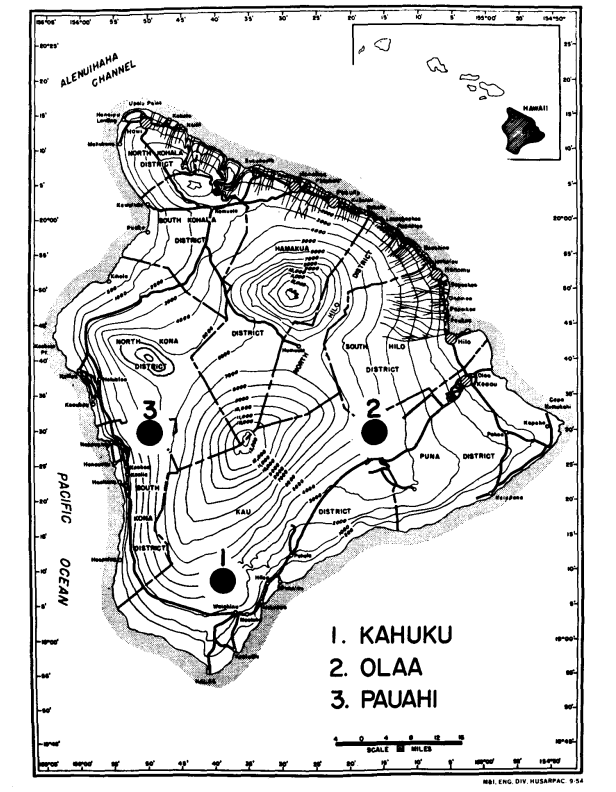
After removal, the abdomen was frozen overnight and then individually homogenized in 0.05 ml deionized distilled water and each supernatant absorbed by four wicks of Whatman No. I filter paper (1 X 0.2 cm). These wicks were then separated and applied to four horizontal starch gels combining different buffer systems. Following electrophoresis, each gel was sliced horizontally four or five times, and these slices stained separately for one or more of 25 enzymes. All work was done using Electrostarch lot 303 at 12% Concentration.
The three buffer combinations used in the study are as follows: Buffer System A = Gel buffer: pH 8.9, 0.0076 m Tris, 0.005 m citrate; Electrodes buffer: pH 8.7, 0.269 m borate, 0.1 m sodium hydroxide. Buffer System C = Gel buffer: pH 8.1, 0.074 m Tris, 0.008 m citrate; Electrodes buffer: pH 8.1, cathode = 0.343 m Tris, 0.079 m citrate; anode = 0.458 m Tris, 0.0104 m citrate. Buffer System JRP = Gel buffer: pH 7.0, 0.009 m Tris, 0.003 m citrate; electrode buffer: 0.135 m Tris, 0.040 m citrate. All these buffer and the staining methods used are similar to those described by Steiner and Johnson (1973) and Ayala et al. (1972; 1974a).
The alleles at a particular locus were numbered according to the relative mobilities of their allozymes on the specified electrophoretic buffer systems. The allele producing the most frequent allozyme was designated l.00 and alleles corresponding to faster or more slowly migrating bands were arbitrarily assigned values indicative of their respective mobilities.
The buffer used and the notation for each enzyme is following: Buffer System A : Esterase (EST, 1 locus) ; Alkaline Phosphatase (AP, 1 locus) ; Acid Phosphatase (ACPH, 1 locus) ; Glutamate Oxaloacetate Transaminase (GOT, 2 loci) ; Aldehyde Oxidase (AO, 1 locus) ; Octanol dehydrogenase (ODH, I locus) ; Alcohol Dehydrogenase (ADH, 1 locus) ; Xantine Dehydrogenase (XDH, 1 locus) ; Leucine Aminopeptidase (LAP, 2 loci) ; Glyceraldehyde-3-Phosphate Dehydrogenase (G-3-PDH, 1 locus). Buffer System C : Malic Enzyme (ME, 1 locus) ; Malate Dehydrogenase (MDH, 2 loci) ; α-Glycerophosphate Dehydrogenase (α-GPDH, 1 locus) ; Phosphoglucomutase (PGM, I locus) ; Fumarase (FUM, 1 locus) ; Sorbitol Dehydrogenase (SDH, 1 locus) ; Hexokinase (HK, 2 loci) ; Isocitrate Dehydrogenase (IDH, 1 locus). Buffer System JRP : 6-Phosphogluconic Dehydrogenase (6-PGDH, 1 locus) ; Hydroxybutyrate Dehydrogenase (HBDH, 1 locus) ; Phosphoglucose Isomerase (PGI, 1 locus).
Results
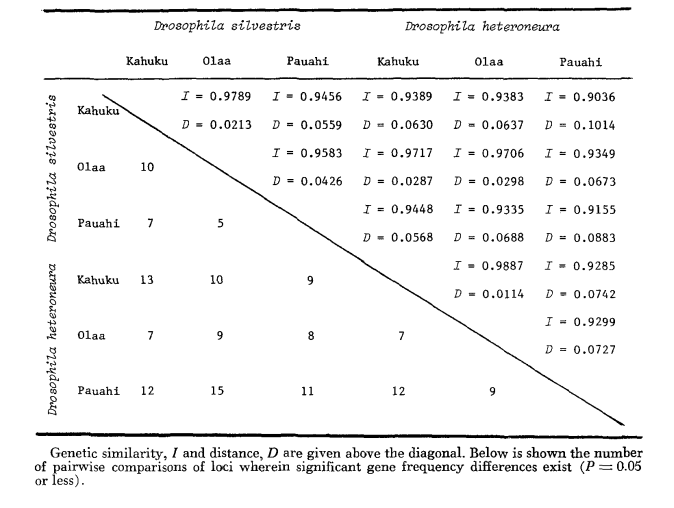
The basic allozyme data are given in Table 1. Table 2 shows the results of using the data in Table 1 to calculate coefficients of genetic similarity, I, and distance, D (Nei 1972) in a pairwise manner within and between the species. In the same table (below the diagonal) are shown the number of loci which display differences in gene frequency (P ≤ 0.05) in pairwise comparisons. The calculation was made, using χ2, by using the frequency of the more common allele against a pooled frequency of the other alleles. Yates’ correction was used.
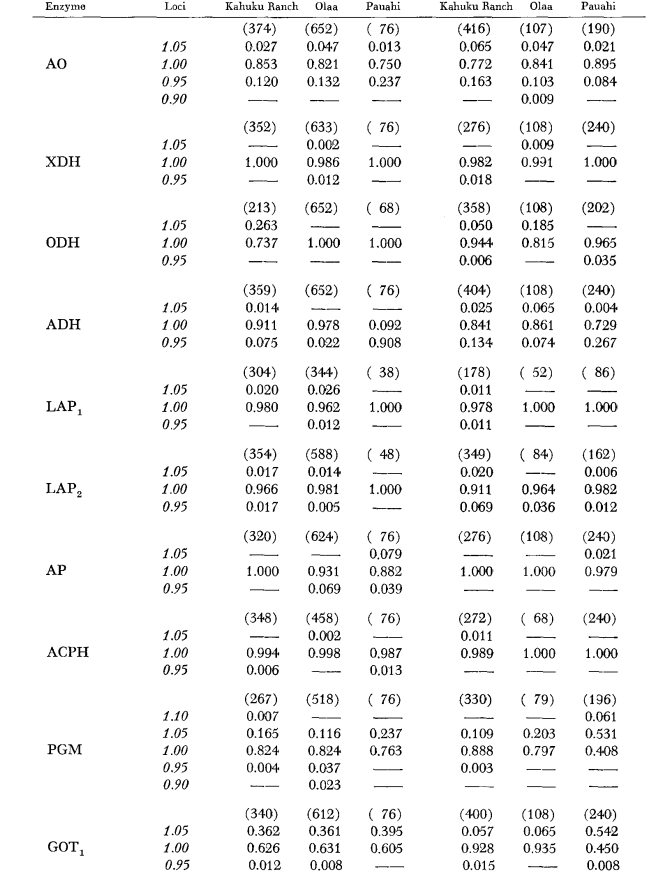
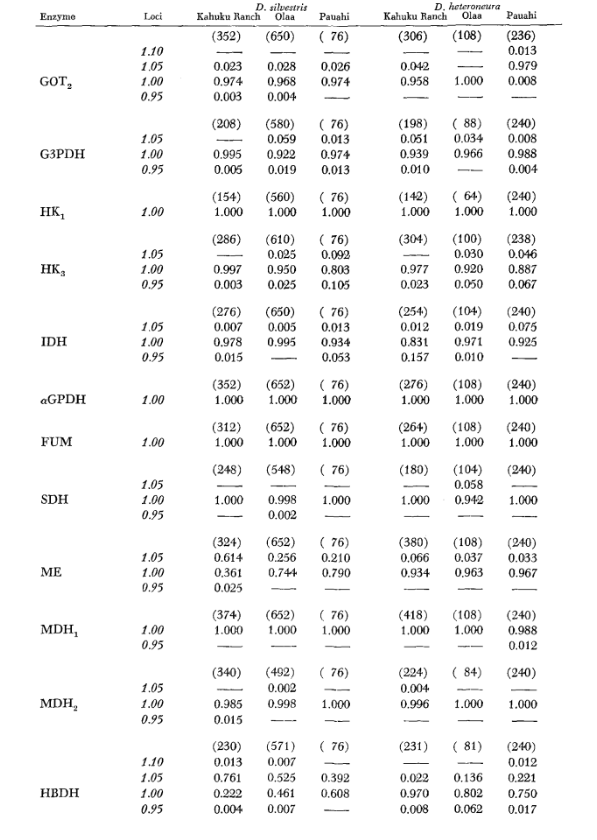
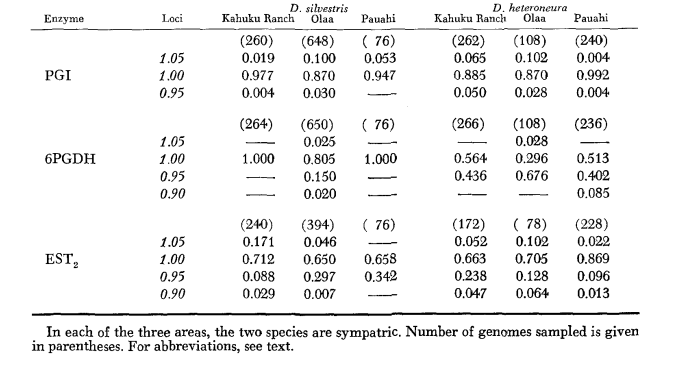
Average heterozygosity levels and estimates of the frequency of polymorphic loci are given in Table 3. A graphic representation of the frequency of allele 1.0 for each allozyme in each population is presented in Figure 2.
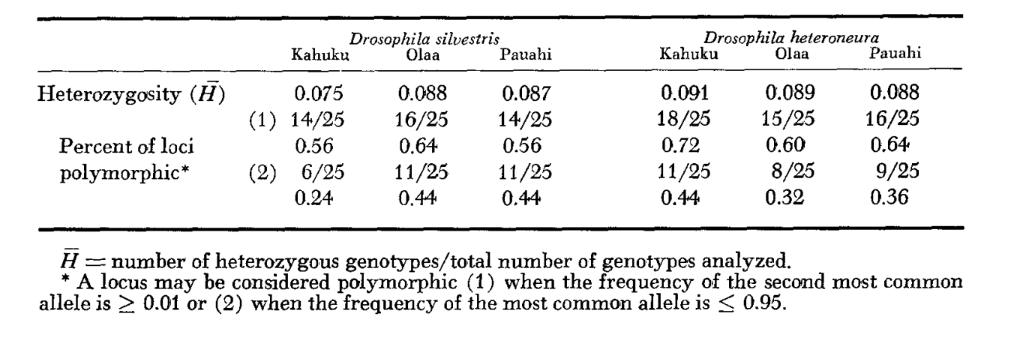
Allozymic similarity is very great both within and between the species, whether measured by similarity coefficients or by differences in gene frequency (see summary in Table 4). There are no fixed allelic differences either within or between the species. At the three areas of sympatry the species show gene frequency differences (P = 0.05) at 9, 11 and 13 loci respectively. This is not much different from the variation within either one of the species across the three localities (silvestris: 5, 7, 10; heteroneura: 7, 9, 12). The two species have similar heterozygosity levels (silvestris 0.083; heteroneura 0.089) and percent of poly- morphic loci (both 0.37).

Some details are of interest. Similarity between the species is least at Pauahi (Table 2; Figure 2). Inspection shows this to be principally due to the frequency differences at ADH arid GOT2. Large differences at these loci appear between the species only in this area; that is, the 1.00 GOT2 allele is low only in Pauahi heteroneura and the 1.00 allele of ADH is low only in Pauahi silvestris (Table 1; Figure 2).
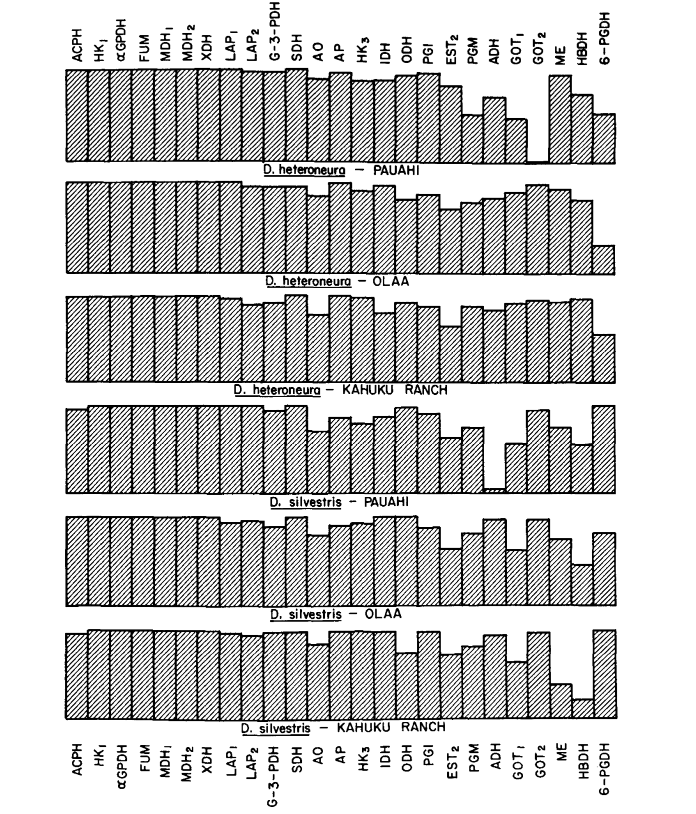
Similarity between the species is somewhat greater in the Kahuku area. Here, the differences involve mainly GOT1, ME arid HBDH. At Olaa the similarity between the species is greatest. GOT1 and HBDH again show differences, although they are not quite as great as those between the species at Kahuku.
Discussion
Genetic comparisons of populations using allozymes have flooded the literature in the last ten years (see reviews by Ayala 1975 and Powell 1975). Since both the proteins studied and the electrophoretic methods used have been, on the whole, similar, findings from quite disparate taxa may be usefully compared. For example, genetic similarity between two species of lizards, when reduced to a coefficient such as that of Nei, may be directly compared with similar data from species pairs of mice, plants or Drosophila.
In their seminal paper, Lewontin and Hubby (1966) pointed out a number of reasons why the standard electrophoretic methods will be biased towards an underestimation of genetic variability and genetic difference. This must be borne in mind all the more today because of the recent development of sequential electrophoretic analysis. Thus Singh, Lewontin and Felton (1976) and Coyne (1976) have shown that, for the Xanthine dehydrogenase locus of D. pseudo-obscura and persimilis, the number of alleles in rather small natural samples of these species is at least four times larger than the number revealed by standard methods. As far as interpopulational comparisons go, these new methods seem certain to reduce the observed similarities at all levels of population differentiation (e.g., local populations, subspecies, semispecies, species).
Despite these new developments, comparisons based on the standard methods continue to be valuable if their limitations are realized. D. silvestris and heteroneura are far closer allozymically than any other Drosophila species-pairs yet examined, including all the pairs of “sibling” species, subspecies and semispecies (Ayala et al. 1974b). Indeed, the similarity coefficients for populations within either silvestris or heteroneura are of the same order of magnitude as those between the same two species. The small differences that do exist between pairs of populations of the two species, furthermore, are not the same in the three areas studied; that is, they do not involve the same loci or alleles.
As discussed in the introduction, we continue to recognize these two entities as good biological species. They are morphologically distinct, widely sympatric, behaviorally isolated and different in their chromosomal polymorphisms. The morphological differences depend on approximately fourteen polygenes (Val 1976).
A striking and important feature of these species, however, is the fact that reciprocal hybrids, obtained in the laboratory, are fully fertile in both sexes (Craddock 1974; Ahearn and Val 1975). Accordingly, natural hybridization must be considered. Although no evidence for this was found by Craddock (1974), Kaneshiro find Val (1977) have reported that approximately two per- cent of the wild flies captured at Kahuku Ranch may be identified as hybrids by morphological means. This is the same region where natural hybrids between two other species were found (Carson, Nair and Sene 1975). Morphometric analysis of head shape has been used with success to specify the exact reciprocal parentage of certain F, hybrid individuals. In one instance, a wild-caught D. silvestris female produced progeny consisting entirely of F, hybrids. Because each wild female was isolated immediately from all males at capture, it is clear that the relevant matings took place in nature. It should be stressed that the background of extensive morphometric studies of laboratory hybrids (Val 1976) has set the stage for these newer natural observations and, accordingly, diagnosis of these cases rests on firm comparative evidence. All natural hybrid individuals have been excluded from the electrophoretic data presented in this paper.
A still further fact which adds to the unusual situation regarding these species is their close ecological similarity. Both have been reared from the same individual decaying stems of the common host plant, Clermontia. Both, likewise, form their leks in very similar places, so that males and females of both species interact in nature.
What is the reason for the very great allozymic similarity of these species? Two possibilities suggest themselves. First, when divergence between the species originally occurred, it may have been accompanied by a certain amount of electrophoretic divergence. The currently observed similarities would thus require that such differences be eroded by introgressive hybridization subsequent to speciation. A second view holds that only minor electrophoretic change accompanied the speciational events in the first place. This might be correlated with the very considerable ecological closeness.
We favor the second view for a number of reasons. The two species are most similar at Olaa. Nevertheless, strong evidence for recent hybridization has been obtained only for Kahuku, where the species are most distant allozymically. At Kahuku, furthermore, the natural populations give no evidence of forming a “hybrid swarm” resembling the broad genetic recombination observed in laboratory crosses. Indeed, introgression between the species is probably occurring, but it may be restricted to a narrow genotypic spectrum by natural selection. Thus only genes of biologically minor import (the “open system,” Carson 1975) may be able to flow across the species barrier. Conversely, other genes may mark or be a functional part of a closed system of epistatic balances unique to one or other of the species. Selection might thus serve to inhibit breakup of such a “closed system” (Carson 1975).
Because of the geological newness of the island of Hawaii, we are tempted to suggest that D, silvestris and D. heteroneura may truly be newly formed in time. This circumstance might be the primary cause of the electrophoretic similarity of the two species. Indeed, they may be far newer than classical “sibling” species pairs (see Carson 1976b). As in other such apparent neospecies, (e.g. Gottlieb 1974; Turner 1974; Avise, Smith and Ayala 1975), allozymic differentiation has been relatively slight. Indeed, the possibility exists that allozymic difference may simply correlate with time elapsed since the cladistic event separated the two compared entities (Avise, Smith and Ayala 1975; Carson 1976a). Morphological differences, on the other hand, may serve as better indicators of the underlying effects of regulatory genes characterizing the important genetic differences between newly formed species.
Authors: F. M. Sene, H. L. Carson
Literature cited:
- Ahearn, J. N., H. L. Carson, Th. Dobzhansky and K. Y. Kaneshiro, 1974 Ethological isolation among three species of the planitibia subgroup of Hawaiian Drosophila. Proc. Natl. Acad. Sci. U.S. 71: 901—903.
- Ahearn, J. N. and F. C. Val, 1975 Fertile interspecific hybrids of two sympatric Hawaiian Drosophila. Genetics 80: s9.
- Avise, J. C., J. J. Smith and F. J. Ayala, 1975 Adaptive differentiation with little genic change between two native California minnows. Evolution '29: 411—426.
- AYALA, F. J., 1975 Genetic differentiation during the speciation process. Evolutionary Biology 8: 1—78.
- Ayala, F. J., J. R. Powell, M. L. Tracey, C. A. Mourão and S. Pérez-Salas, 1972 Enzyme variability in the Drosophila willistoni group. IY Genic variation in natural populations Of Drosophila willistoni. Genetics 70: 113—139.
- Ayala, F. J., M. L. Tracey, L. G. Barr and J. G. Ehrenfeld, l9f4a Genetic and reproductive differentiation of the subspecies, Drosophila equinoxialis caribbensis. Evolution 28: 24—41.
- Ayala F. J., M. L. Tracey, D. Hedgecock and R. C. Richmond, 1974b Genetic differentiation during the speciation process in Drosophila. Evolution 28: 576—592.
- Carson, H. L. 1975 The genetics of speciation at the diploid level. Am. Naturalist 109: 83—92, 1976a Inference of the time of origin of some Drosophila species. Nature 259: 395—396, 1976b Genetic differences between newly-formed species. Bioscience 26: 700—701.
- Carson, H. L., P. S. Nair and F. M. Sene, 1975 Drosophila hybrids in nature: Proof off gene exchange between sympatric species. Science 189: 806—807.
- Carson, H. L. and H. D. Stalker, 1968 Polytene chromosome relationships in Hawaiian species of Drosophila. II. The D. planitibia subgroup. Univ. Texas Publ. 6818: 355-365.
- Coyne, J. A., 1976 Lack of genic similarity between two sibling species of Drosophila as revealed by varied techniques. Genetics 84: 593—607.
- Craddock, E. M., 1974 Reproductive relationships between homosequential species off Hawaiian Drosophila. Evolution 28: 593—606.
- Craddock, E. M. and H. L. Carson, 1975 Chromosome variability in an endemic Hawaiian Drosophila species. Genetics 80: s23.
- Gottlieb, L. D., 1974 Genetic confirmation of the origin of Clarkia lingulata. Evolution 2B: 244-250.
- Hardy, D. E., 1965 insects of Hawaii, Vol. 12. p. 814. University of Hawaii Press, Honolulu.
- Johnson, W. E. and H. L. Carson, 19f5 Allozymic variation in Drosophila silvestris. Genetics 80: s46.
- Johnson, W. E., H. L. Carson, K. Y. Kaneshiro, W. W. M. Steiner and M. M. Cooper, 1975 Genetic variation in Hawaiian Drosophila. II. Allozymic differentiation in the D. planitibia subgroup. pp. 563—584. In: Isozymes IV. Genetics and Evolution. Edited by C. L. Markert. Academic Press, New York.
- Kaneshiro, K. Y., 1976 Ethological isolation and phylogeny in the planitibia subgroup of Hawaiian Drosophila. Evolution 30: 740-745.
- Kaneshiro, K. Y. and F. C. Val, l977 Natural hybridization between a sympatric pair of Hawaiian Drosophila. Am. Naturalist (in press).
- Lewontin, R. C. and J. L. Hubby, 1966 A molecular approach to the study of genic hetero- zygosity in natural populations. II. Amount of variation and degree of heterozygosity in natural populations of Drosophila pseudoobscura. Genetics 54: 595—609.
- Nei, M., 1972 Genetic distance between populations. Am. Naturalist 106: 283—292.
- Powell, I. R., 1975 Protein variation in natural populations of animals. Evol. Biol. 8: 79-113.
- Simon, R. S., R. C. LEWONTin and A. A. Fzczou, 19 Z6 Genetic heterogeneity within electro- phoretic “alleles” of xanthine dehydrogenase in Drosophila pseudoobscura. Genetics 84: 609—629.
- Steiner, W. W. M. and W. E. Johnson, 1973 Techniques for electrophoresis of Hawaiian Drosophila. US/IBP Island Ecosystems IRP. Tech. Report 30: ii + 21 pp.
- Turner B. J., 1974 Genetic divergence of Death Valley pupfish species: Biochemical versus morphological evidence. Evolution fl8: 281-294.
- Val, F. C., 1976 Genetics of morphological differences between two interfertile species of Drosophila. Genetics 83: s78.

